
Best experienced using the browsers Chrome  or Firefox
or Firefox  .
.
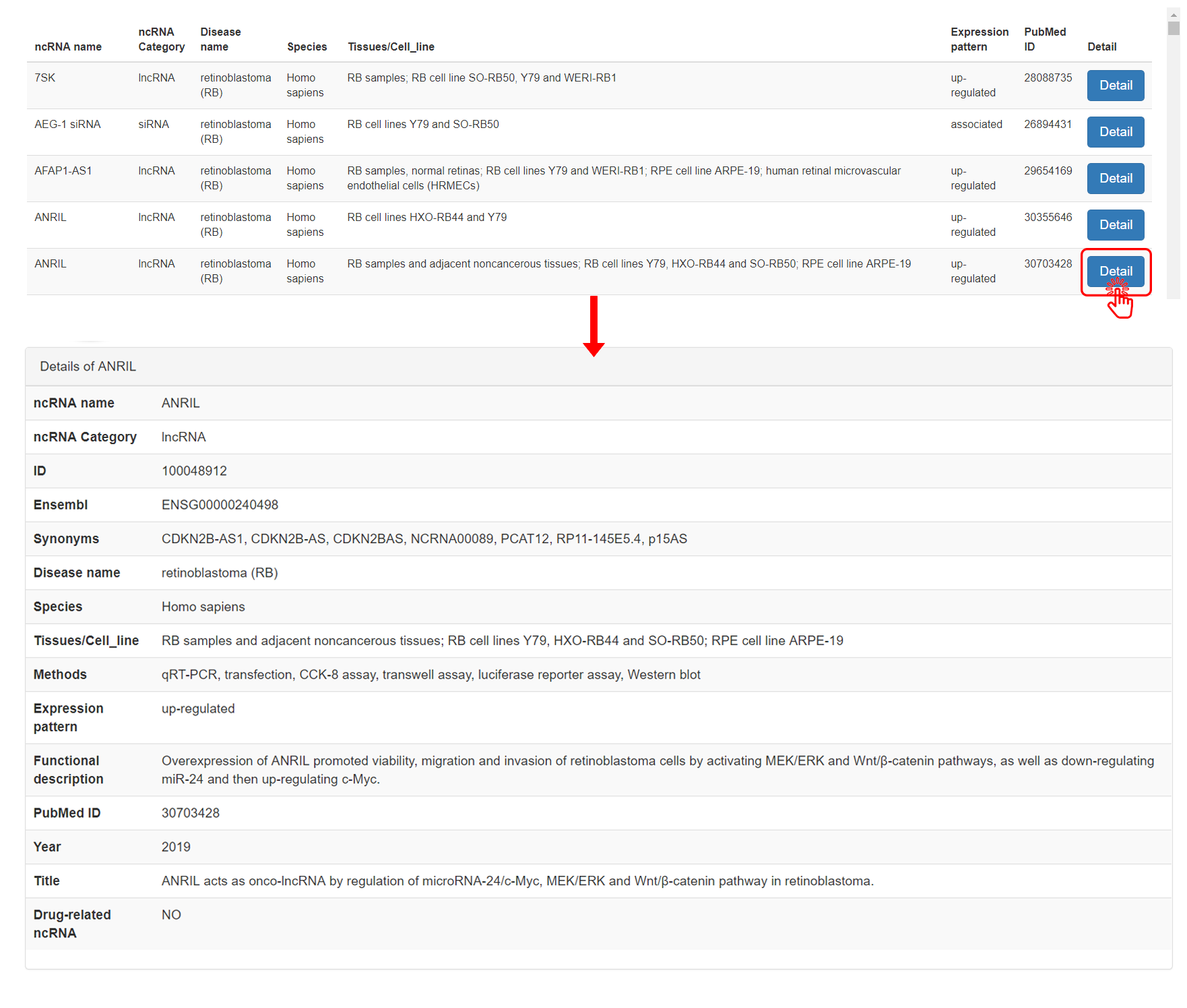
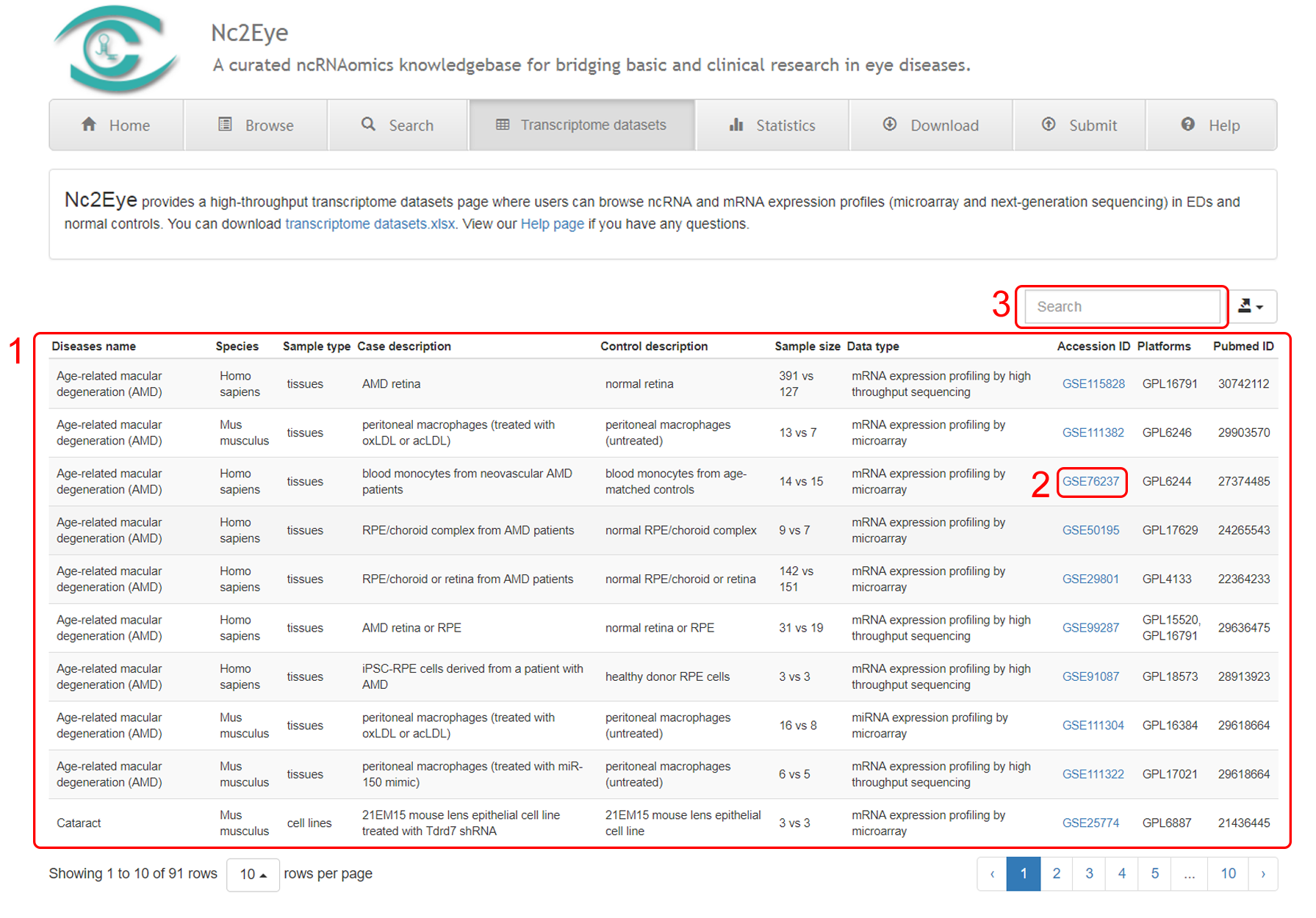
Copyright © Institution of Biomedical Big Data, Wenzhou Medical University All rights reserved.